Microbiomes: DADA2
I like to analyse all of my microbiome data directly in R with the DADA2 package. This is an Amplicon Sequence Variant (ASV) approach (meaning reads aren't clustered according to a similarity threshold) and relies on run-specific error models to denoise data and limit overestimations of sequence diversity. The DADA2 tutorial is a great place to start if you're interested in this method. Alternatively, you can implement DADA2 in QIIME2.
Metagenomes and MAGs: Anvio
Anvio is an immense resource for analysing metagenomes and metagenome-assembled genomes (MAGs). It has a brilliant graphical interface that can be used to manually refine bins but Anvio also wraps a whole suite of tools for pangenomics, phylogenomics, etc. Their webpages include a number of highly detailed tutorials to help you think about your own data and the visualisations are always superb.
Eukaryotic Phylogenomics: Phylofisher
Phylofisher is a very handy collection of scripts and tools for eukaryotic phylogenomics (i.e. evolutionary histories inferred from multiple genes/proteins). It involves manually inspecting single-gene trees prior to concatenation. Using their precompiled database, you can generate a set of 240 single-gene trees which include 304 reference eukaryotes. To remove contaminants or identity paralogs in single-gene trees, they also provide a very nice application called Parasorter which makes the more labour intensive part of the process much more manageable.
Happy Belly Bioinformatics
This is an excellent resource from Mike Lee and one I often share with those who are new to bioinformatics. It also includes tutorials for amplicon analysis and genomics using some of the methods above.
PR2
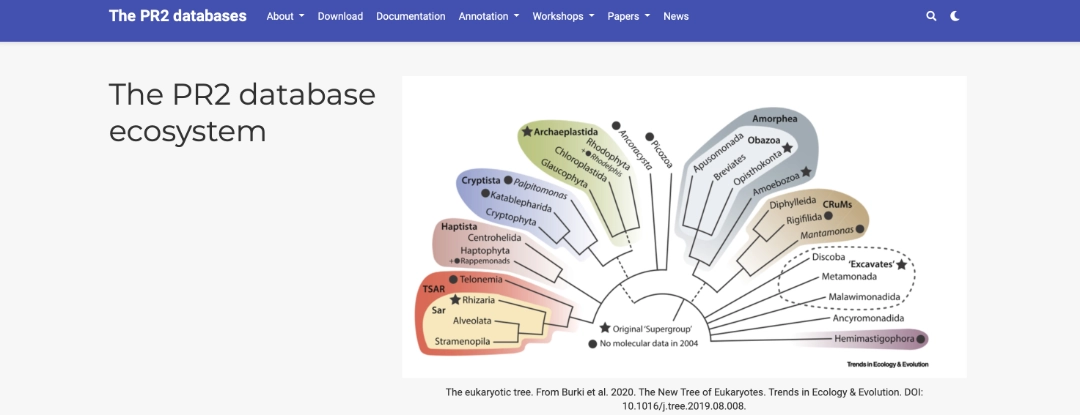
The PR2 or Protist Ribosomal Reference database is a curated collection of eukaryotic SSU sequences. It is regularly updated and refined, and is now my go-to for any protist microbiome work. I'd also recommend the pr2 R package listed below.
R packages
tidyverse
A collection of R packages designed for data science.
phyloseq
Tools to import, store, analyse, and graphically display microbiome census data.
vegan
Community ecology tools for diversity analysis, ordination, and other functions for ecologists.
ggtree
Visualisation and annotation of phylogenetic trees.
ggtreeExtra
Extensions for ggtree, providing additional functionalities for circular tree visualisation.
ape
Functions for reading, writing, plotting, and manipulating phylogenetic trees.
microbiome
Analysis and visualisation of microbiome data.
igraph
Network analysis and visualisation.
treeio
Reading and writing phylogenetic tree in various formats.
patchwork
Organising plots into multi-panelled figures.
DESEQ2
Differential abundance/expression analysis.
ANCOMBC
Differential abundance analysis with bias correction.
RColorBrewer
ColorBrewer palettes and extrapolation.
prismatic
Colour manipulation.
chroma
Colour manipulation and Plotting in R.
pheatmap
Pretty heatmaps.
SpiecEasi
SParse InversE Covariance estimation for Ecological Association and Statistical Inference.
ggridges
Ridgeline Plots in ggplot2.
Biostrings
Efficient manipulation of biological strings.
pr2
PR2 database interface for eukaryotic taxonomy.